Sentinels of Coastal Ecosystem Health
The sensitivity of an aquatic organism's physiological response to environmental change can be leveraged to gain more insight into ecosystem-level processes. The clam, Macoma balthica, is a climate-sensitive species that is found from temperate to Arctic latitudes in both the Atlantic and the Pacific. In the summer of 2014 we collected M. balthica from Puget Sound, WA with our high school intern, Anastasia, and Brook's daughter, Tasman. We spent a few hours digging for clams with the goal of collecting enough to do exposures to elevated temperature, shellfish bacterial pathogens, and temperature + bacterial pathogens. After a lab acclimation, the clams were subjected to a 24-hour exposure to one of the environmental "stress" treatments or were maintained in control conditions. We explored the proteomic responses of these clams to the different environmental conditions to better understand the mechanisms behind M. balthica's documented sensitivity to environmental change.
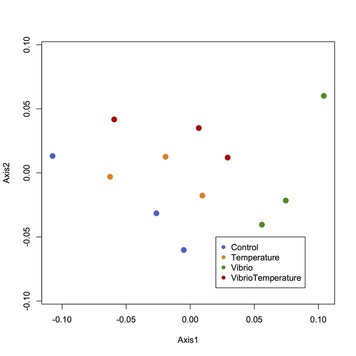
We identified 2848 proteins in the M. balthica proteome after sequencing on the Q-Exactive mass spectrometer. The figure to the right shows a multivariate analysis of clam proteomic response to the different environmental treatments. Each dot represents a clam and clams that are closer together have more similar proteomic profiles. More than 300 proteins were differentially abundant in the bacteria exposure (green) and the bacteria + temperature exposure (red) clams when compared to clams kept in control conditions (blue). More than 100 proteins were differentially abundant in the elevated temperature-exposed clams (green). From these results we can begin to uncover why the clams are sensitive to these environmental changes.


