Bacteria Recyclers in the Ocean
Jessica Faux- Ph.D thesis project
A collaboration between the Harvey and Nunn lab
Potential changes in global climate are likely to alter biogeochemical cycles around the world. We are interested in understanding more about the potential impacts of climate change on organic carbon and nitrogen cycling in marine environments. Before an examination of climate impacts can be attempted we need to more fully understand all components of these cycles. The role of proteins and their ultimate fate in marine systems is not fully understood. Proteins are labile and readily recycled but we don’t have a complete understanding of how proteins make their way through marine systems or how much proteinaceous material is eventually preserved in sediments. With the ultimate goal of expanding our knowledge of the role of proteins in marine systems we began by investigating what proteins are produced through the growth phases of a model bacterium. Bacteria are essential for the cycling and degradation of organic matter. It is because of their pivotal role that we chose to start by examining what proteins they produce.
|
In addition to studying the total proteins produced by this bacterium we were also interested in whether we could track the production of proteins produced by giving the bacterium a labeled substrate. A second investigation was undertaken in which the bacterium was given a labeled amino acid and its incorporation into proteins tracked. We identified over 2000 proteins produced by this bacterium which accounts for 48% of the theoretical proteome (Fig. 1). We also successfully tracked the incorporation of the labeled amino acid into the proteins produced by the model bacterium (Fig. 2). This demonstrates a method by which protein production can be tracked allowing for further investigation into the production, recycling, and degradation of proteins within marine systems offering the possibility of learning more about their role in marine systems.
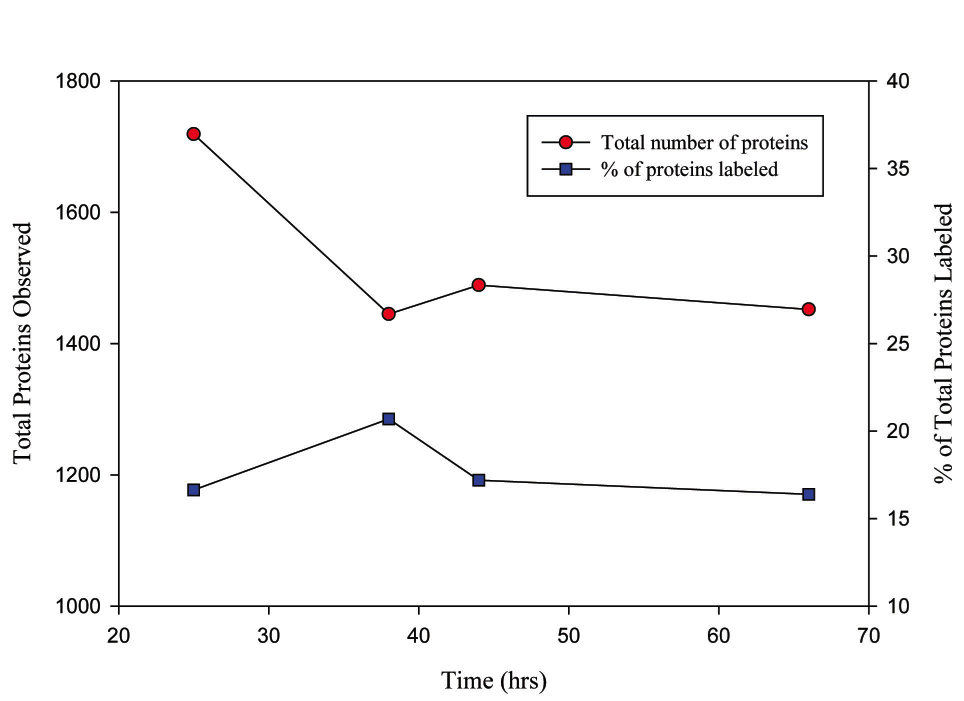
Figure 2. Sum of R. pomeroyi proteins observed and the percentage of that total that was labeled with 13C-leucine in all replicates at the four time point plotted by hours. The lists of proteins found in the three replicates for each time point were combined and duplicate, or over-lapping, proteins were deleted to compile a list of all the proteins seen within a time point. Proteins were defined as having match probability of 90% or greater by ProteinProphet and PeptideProphet and having 2 or more independent spectra identified for the protein.
|
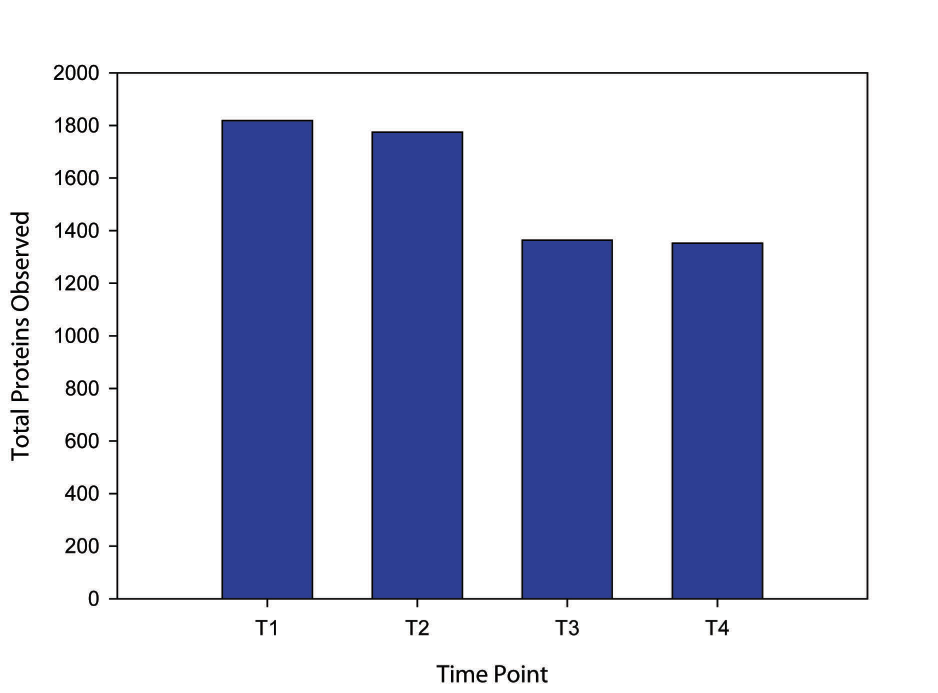
Figure 1. Sum of R. pomeroyi proteins observed in all replicates of a time point. The lists of proteins found in the three replicates for each time point were combined and duplicate, or over-lapping, proteins were deleted to compile a list of all the proteins seen at each time point. Proteins were defined as having match probability of 90% or greater by ProteinProphet and PetptideProphet and having 2 or more independent spectra identified for the protein.
|
Results and Discussion
We examined the total expressed proteome of the heterotrophic marine bacterium, Ruegeria pomeroyi under environmentally realistic carbon conditions. We tracked the proteins produced at four time points representing early exponential growth phase, late exponential growth phase, early stationary phase and late stationary phase: 25, 38, 44 and 66 hours, respectively. We identified 2037 proteins throughout the four growth phase that equated to 48% of the theoretical proteome. The total number of observed proteins decreased over time (Fig.1) as did the number of proteins common between adjacent time points. Despite such decreases, at least 75% of proteins in common among adjacent time points remained. The most abundant proteins identified were responsible for porins, transport, binding, translation, and protein refolding. An examination of the functional annotation of expressed proteins found clusters most enriched within the samples contained proteins that contributed to RNA subunit structure and RNA binding; ribosome assembly and biogenesis; translation and amino acid, amine, organic acid, and oxoacid biosynthesis and metabolism. The biological functions identified among the most abundant proteins and the most prevalent clusters suggest an organism that maintained active growth well into stationary phase with no obvious patterns of changing biological function. The presence of abundant porins and transport proteins relative to the other proteins produced could represent potential proteins to be used as biomarkers of bacterial processes and/or activity.
A second investigation using R. pomeroyi, was conducted in which this bacterium was cultured with a tracer level addition of 13C-labeled leucine (average addition: 600-800nM). Bacterial protein biosynthesis which incorporated the label was tracked through exponential and stationary growth phases (same sampling time points as listed above). Over 1900 total proteins were identified, with a total of 479 proteins incorporating the isotopic leucine label. Label incorporation ranged between 16 and 21% of the total proteins depending on growth phase (fig. 2). The widespread distribution of the label among the time points indicates that the isotopic label was recycled by the bacteria with little remineralization (Fig.3). This study demonstrates the successful tracking of the uptake and incorporation of an isotopically labeled amino acid and demonstrates a method by which bacterial protein synthesis can be tracked.
A second investigation using R. pomeroyi, was conducted in which this bacterium was cultured with a tracer level addition of 13C-labeled leucine (average addition: 600-800nM). Bacterial protein biosynthesis which incorporated the label was tracked through exponential and stationary growth phases (same sampling time points as listed above). Over 1900 total proteins were identified, with a total of 479 proteins incorporating the isotopic leucine label. Label incorporation ranged between 16 and 21% of the total proteins depending on growth phase (fig. 2). The widespread distribution of the label among the time points indicates that the isotopic label was recycled by the bacteria with little remineralization (Fig.3). This study demonstrates the successful tracking of the uptake and incorporation of an isotopically labeled amino acid and demonstrates a method by which bacterial protein synthesis can be tracked.