PROTEOMIC SERVICES
The most cost effective way to do proteomics is to collaborate with proteomics experts.
Our research focuses on marine proteomics, ocean proteomics, metaproteomics, non-model proteomics, and extreme environment proteomics. We have completed studies on a range of aquatic environments including sediments, bacteria, phytoplankton, and marine invertebrates.
We offer a variety of services. Please email us with:
- A description of your project
- Questions you are trying to answer
- Types of samples you have
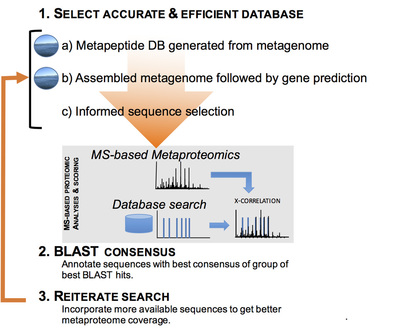
- Information on accompanying genome or metagenome
We can help guide your graduate students or postdoctoral fellows through their analyses.
We have been working in the field of environmental proteomics for over 15 years. We exclusively use Thermo Fisher mass spectrometers with inline Waters NanoAcquity HPLCs, providing us with state-of-the art chromatographic separation and high mass accuracy on all peptides analyzed.
Our lab has access to over 10 Thermo Fisher mass spectrometers dedicated to proteomics experiments and maintained full time by senior scientists.
Our lab has access to over 10 Thermo Fisher mass spectrometers dedicated to proteomics experiments and maintained full time by senior scientists.
COST CENTER
We decided to establish a cost center because so many PIs were discovering the power of proteomics and our lab's abilities after grants were funded. This provides us with an avenue to work together, even if the funding is already in place. We are also seeking new, long-term collaborations in the fields of ocean and environmental proteomics.
ESTIMATE: A back-of-the-envelope calculation for analyzing 1 sample on an Orbitrap based instrument (i.e., the QExactive, or OT) is ~$260 per sample injection/analysis (assuming a 90 minute gradient or 2 hour run time). The cost to analyze samples depends on the experimental design (DIA vs DDA vs targeted analyses), instrument choice (QE, Fusion, Lumos, Velos..). Downstream bioinformatic analyses tend to be more time consuming than people anticipate. As a result, this is charged with a specific rate based on the project. Basic bioinformatics completed on well-annotated model organisms is included in the above estimate.
EXAMPLES OF WORK WE HAVE DONE:

Additionally, there is the the University of Washington Mass Spectrometry Center in the UW Health Sciences Center is open-access self-sustaining instrument laboratory providing a wide variety of mass spectral services to the University and the research community-at-large (including commercial) on a recharge basis. Examples of routine analysis are: Low and High Resolution mass analysis, metabolomics, analyte quantitation, and structural elucidation.
Please note that we are looking for collaborations- we are not a typical cost center "drop off" service. Further, we reserve the right to turn down projects based on our schedule.