Miranda MudgeMiranda is a PhD candidate in the Molecular and Cellular Biology Department working jointly in the MacCoss and Nunn labs. She is broadly interested in using proteomics to better understand how microbiomes influence their environment, specifically by adapting DIA methods for metaproteomics. Her project involves using marine microbiome peptides as biomarkers to forecast the formation of harmful algal blooms. Within the lab, Miranda has also studied how environmentally-relevant cold active bacteria maintain cellular activity after long-term exposure to extreme conditions, as well as how microbiome controls on eukaryotes inhibit viral infections.
Prior to joining the lab, Miranda studied DNA damage repair at Washington University in St. Louis, as well as developed a mouse melanoma therapeutic model at Missouri State University, where she graduated with a M.S. in Cellular and Molecular Biology. |
Research Experience
2018 - present Molecular and Cellular Biology Ph.D. program, University of Washington, MacCoss and Nunn Labs
2015 - 2018 Laboratory technician/manager, Washington University in St. Louis, Mosammaparast Lab
2014 - 2015 Graduate assistant, Laboratory instructor - Biomolecular Interactions, Missouri State University
2013 - 2015 Graduate assistant, Laboratory instructor - Introduction to Biomedical Sciences, Missouri State University
2013 - 2014 Cellular and Molecular Biology M.S. program, Missouri State University, DeLong Lab
2009 - 2013 Cellular and Molecular Biology B.S. program, Missouri State University
2015 - 2018 Laboratory technician/manager, Washington University in St. Louis, Mosammaparast Lab
2014 - 2015 Graduate assistant, Laboratory instructor - Biomolecular Interactions, Missouri State University
2013 - 2015 Graduate assistant, Laboratory instructor - Introduction to Biomedical Sciences, Missouri State University
2013 - 2014 Cellular and Molecular Biology M.S. program, Missouri State University, DeLong Lab
2009 - 2013 Cellular and Molecular Biology B.S. program, Missouri State University
Publications
- Mudge, M.C., Riffle, M., Chebli, G., Plubell, D., Rynearson, T., Noble, W.S., Timmins-Schiffman, E., Kubanek, J., and Nunn., B.L. (2024) Harmful algal blooms preceded by a predictable and quantifiable shift in the oceanic microbiome. Nat. Commun. (in review).
- Nunn., B.L., Timmins-Schiffman, E., Mudge., M.C., et. al. (2024) Microbial metagenomes across a complete phytoplankton bloom cycle: high resolution sampling every 4 hours over 22 days. Scientific Data. (in review).
- Nunn, B.L., Brown, T., Timmins-Schiffman, E., Mudge, M., Riffle, M., Axworthy, J.B., and Dilwort, J. (2024) Resilience in a time of stress: revealing the molecular underpinnings of coral survival following bleaching events. bioRxiv. https://doi.org/10.1101/2024.02.587798.
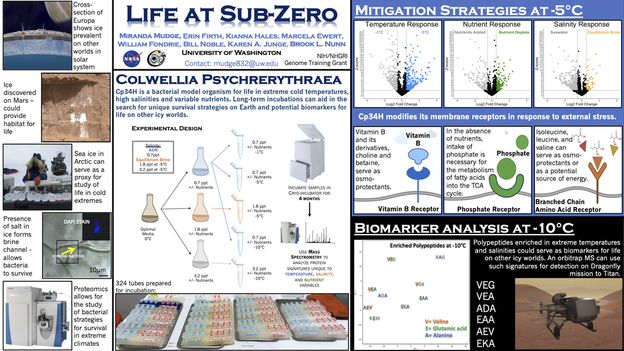
- Mudge, M.C., Nunn, B.L., Firth, E., Ewert, M., Hales, K., Fondrie, W.E., Noble, W.S., Toner, J., Light, B., and Junge, K.A. (2021) Subzero, saline incubations of Colwellia psychrerythraea reveal strategies and biomarkers for sustained life in extreme icy environments. Environmental Microbiology 23, 3840-3866.
- Pollara, S.B., Becker, J.W., Nunn, B.L., Boiteau, R., Repeta, D., Mudge, M.C., Downing, G., Chase, D., Harvey, E.L., and Whalen, K.E. (2021) Bacterial quorum-sensing signal arrests phytoplankton cell division and impacts virus-induced mortality. mSphere 6, e00009-21.
- Byrum, A.K., Carvajal-Maldonado, D., Mudge, M.C., Valle-Garcia, D., Majid, M.C., Patel, R., Sowa, M.E., Gygi, S.P., Harper, J.W., Shi, Y., et al. (2019). Mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function. J Cell Biol 218, 422–432.
- Soll, J.M., Brickner, J.R., Mudge, M.C., and Mosammaparast, N. (2018). RNA ligase-like domain in activating signal cointegrator 1 complex subunit 1 (ASCC1) regulates ASCC complex function during alkylation damage. J Biol Chem 293, 13524–13533.
- Zhao, Y., Mudge, M.C., Soll, J.M., Rodrigues, R.B., Byrum, A.K., Schwarzkopf, E.A., Bradstreet, T.R., Gygi, S.P., Edelson, B.T., and Mosammaparast, N. (2018). OTUD4 Is a Phospho-Activated K63 Deubiquitinase that Regulates MyD88-Dependent Signaling. Mol Cell 69, 505-516.e5.
- Brickner, J.R., Soll, J.M., Lombardi, P.M., Vågbø, C.B., Mudge, M.C., Oyeniran, C., Rabe, R., Jackson, J., Sullender, M.E., Blazosky, E., et al. (2017). A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature 551, 389–393.
- DeLong, R.K., Mitchell, J.A., Morris, R.T., Comer, J., Hurst, M.N., Ghosh, K., Wanekaya, A., Mudge, M., Schaeffer, A., Washington, L.L., et al. (2017). Enzyme and Cancer Cell Selectivity of Nanoparticles: Inhibition of 3D Metastatic Phenotype and Experimental Melanoma by Zinc Oxide. J Biomed Nanotechnol 13, 221–231.
- Ramani, M.*, Mudge, M.C.*, Morris, R.T., Zhang, Y., Warcholek, S.A., Hurst, M.N., Riviere, J.E., and DeLong, R.K. (2017). Zinc Oxide Nanoparticle-Poly I:C RNA Complexes: Implication as Therapeutics against Experimental Melanoma. Mol Pharm 14, 614–625.
Conferences
|
Oral Presentations
Poster Presentations
|
Astrobiology Field Workshop 2019 – Friday Harbor
Fun times outside the lab, featuring my dog Brody