Iron limitation on diatoms
|
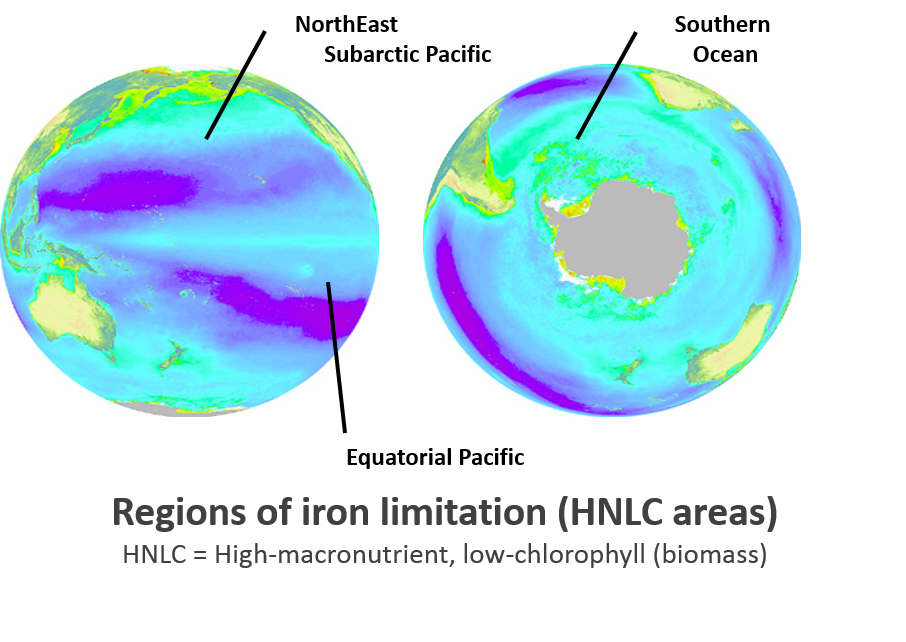
Vast areas of the world’s
oceans are classified High Nutrient Low Chlorophyll (HNLC) regions. Studies in the early 1990’s demonstrated that
iron availability limits growth of photosynthetic organisms. Microscopic photosynthetic organisms in the
ocean, like diatoms, are responsible for > 50% of global O2
production. In addition, because diatoms
have a silica shell (frustule), their
death can provide a rapid and direct means for transporting CO2 from
the atmosphere and upper ocean to the deep ocean.
In the early 90's the idea of iron fertilization in the ocean as a means for restoring plankton blooms was tested by oceanographers. Since then, several start-ups have tried to capitalize on this simplistic view for solving global warming. The biochemistry within these microscopic organisms is far more complex than this simplistic view suggests and requires further investigation to understand the links between blooms and climate change. We investigated Thalassiosira pseudonana, a cosmopolitan diatom, grown under nutrient replete and iron limiting conditions to better understand how diatoms survive in iron-limited waters. This was an exploratory project to try and decipher what metabolic strategies must be altered in order to continue photosynthesis when most of the photosynthetic complexes require iron. Rather than focusing on single proteins or groups of proteins that are involved in on or more metabolic processes, we tried to look at whole cells metabolic transformation. Our work is published in PLOS ONE. |
PLOS ONE: Diatom Proteomics Reveals Unique Acclimation Strategies to Mitigate FE Limitation
Published in PLOS ONE
Nunn BL, Faux JF, Hippmann AA, Maldonado MT, Harvey HR, et al. (2013) Diatom Proteomics Reveals Unique Acclimation Strategies to Mitigate Fe Limitation. PLOS ONE 8(10): e75653.
Nunn BL, Faux JF, Hippmann AA, Maldonado MT, Harvey HR, et al. (2013) Diatom Proteomics Reveals Unique Acclimation Strategies to Mitigate Fe Limitation. PLOS ONE 8(10): e75653.
|
Phytoplankton growth rates are limited by the supply of iron (Fe) in approximately one third of the open ocean, with major implications for carbon dioxide sequestration and carbon (C) biogeochemistry. To date, understanding how alteration of Fe supply changes phytoplankton physiology has focused on traditional metrics such as growth rate, elemental composition, and biophysical measurements such as photosynthetic competence (Fv/Fm). Researchers have subsequently employed transcriptomics to probe relationships between changes in Fe supply and phytoplankton physiology. Recently, studies have investigated longer-term (i.e. following acclimation) responses of phytoplankton to various Fe conditions. In the present study, the coastal diatom, Thalassiosira pseudonana, was acclimated (10 generations) to either low or high Fe conditions, i.e. Fe-limiting and Fe-replete. Quantitative proteomics and a newly developed proteomic profiling technique that identifies low abundance proteins were employed to examine the full complement of expressed proteins and consequently the metabolic pathways utilized by the diatom under the two Fe conditions.
|
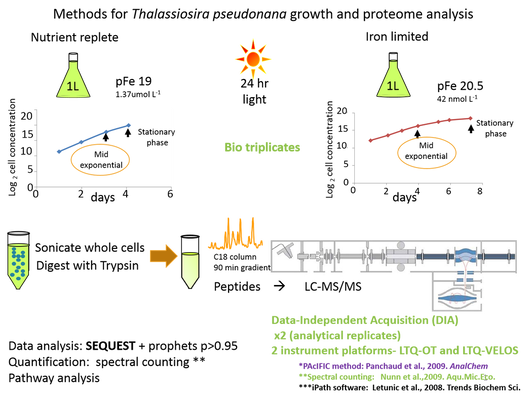
Cartoon representation of the methods used to identify proteomic response of the diatom Thalassiosira pseudonana to growth in iron limitation. Cells were acclimated (10 generateions) and collected at mid-exponential growth phase. Samples were analyzed on the LTQ-OT using a data-dependent method in quadruplicate and on the LTQ-VELOS using a data independent method (PAcIFIC) in triplicate.
|
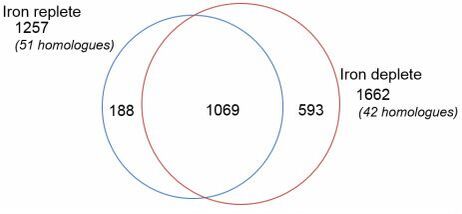
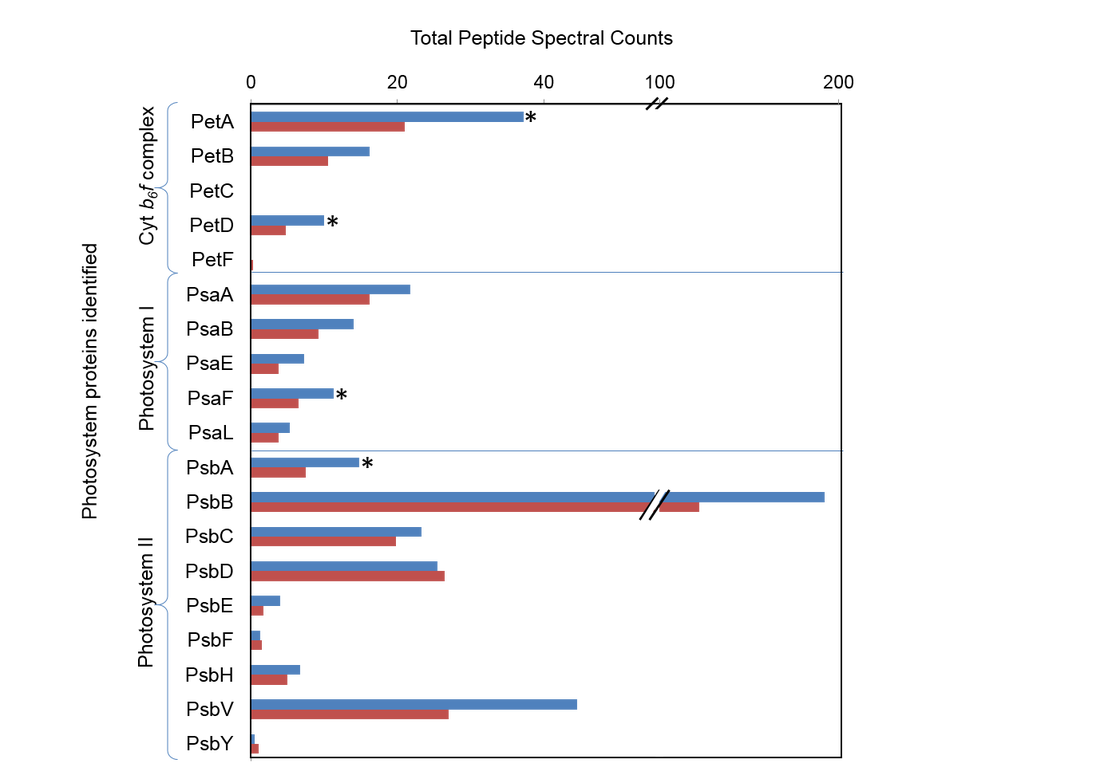
A total of 1850 proteins were confidently identified, nearly tripling previous identifications made from differential expression in diatoms. Given sufficient time to acclimate to Fe limitation, T. pseudonana up-regulates proteins involved in pathways associated with intracellular protein recycling, thereby decreasing dependence on extracellular nitrogen (N), C and Fe. The relative increase in the abundance of photorespiration and pentose phosphate pathway proteins reveal novel metabolic shifts, which create substrates that could support other well-established physiological responses, such as heavily silicified frustules observed for Fe-limited diatoms. Here, we discovered that proteins and hence pathways observed to be down-regulated in short-term Fe starvation studies are constitutively expressed when T. pseudonana is acclimated (i.e., nitrate and nitrite transporters, Photosystem II and Photosystem I complexes). Acclimation of the diatom to the desired Fe conditions and the comprehensive proteomic approach provides a more robust interpretation of this dynamic proteome than previous studies.

Cartoon representation of diatom cell biochemistry when acclimated to Fe-limitation. Not all metabolic pathways are shown. Black and grey pathways and proteins indicate presence during Fe-limitation, white proteins indicate significantly down-regulated proteins, colored pathways were significantly up-regulated in Fe-limited cells compared to Fe-replete cells. A) Pentose Phosphate Pathway, B) The rejoining of the pentose phosphate pathway with glycolysis and generation of pyruvate. C) Polyamine synthesis using spermine synthase. D) Proposed reduction of Fe+3 and eventual transport of Fe into cells. Purple: photosynthesis and glycolysis-gluconeogenesis, red: nucleotide metabolism, teal: lipid metabolism, orange: amino acid metabolism.
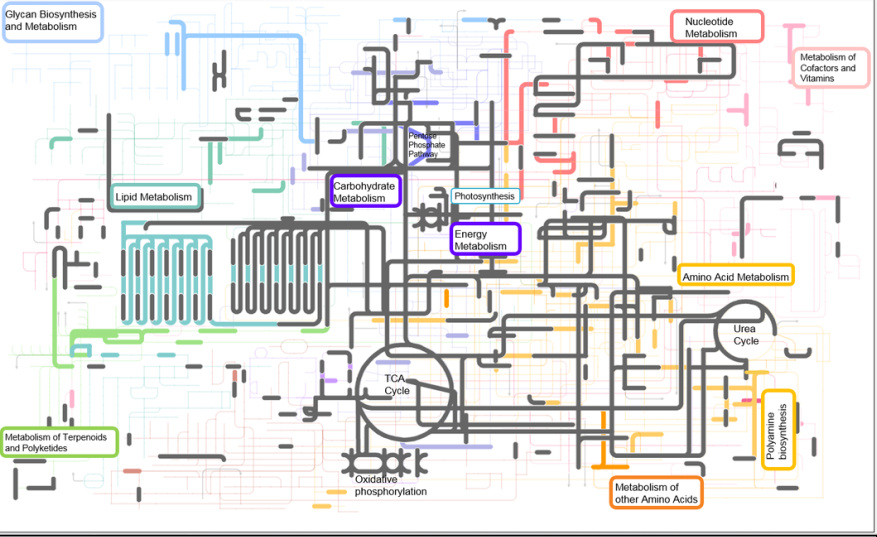
Metabolic biochemistry map of proteins expressed and identified in Fe-limited T. pseudonana. Map includes data from triplicate PAcIFIC analyses on a tandem mass spectrometer from Thalassiosira pseudonana acclimated to Fe-limitation. Each node (or corner) represents a metabolite and the lines connecting the nodes represent an enzyme (i.e. protein). Metabolites were not measured in this study. Proteins that were identified in both Fe-replete and Fe-limited cultures are highlighted in grey. Proteins that were identified to be unique to Fe-limited cultures are indicated in color. From top left – light blue: sugar and glycan biosynthesis, light purple: starch and sucrose metabolism (including photosynthesis, oxidative phosphorylation, carbon fixation), dark purple: glycolysis-gluconeogenesis (including TCA cycle), red: nucleotide metabolism, teal: lipid metabolism, orange: amino acid metabolism (including urea cycle).
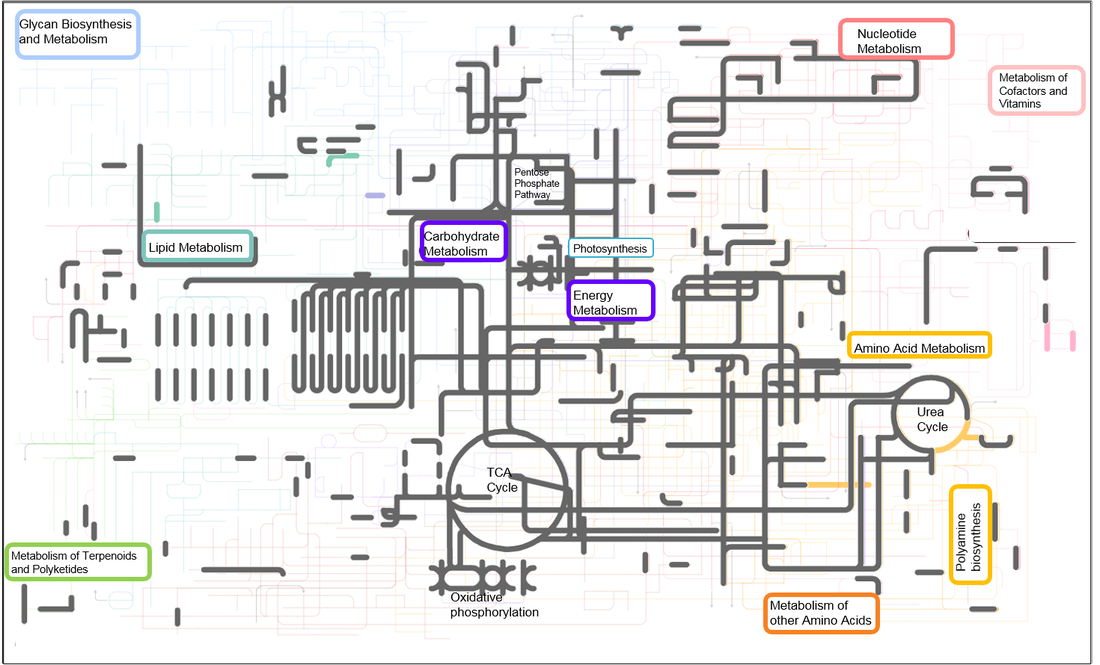
Metabolic biochemistry map of proteins expressed and identified in Fe-replete T. pseudonana. Map includes data from triplicate PAcIFIC analyses on a tandem mass spectrometer from Thalassiosira pseudonana acclimated to Fe-replete conditions. Each node (or corner) represents a metabolite and the lines connecting the nodes represent an enzyme (i.e. protein). Metabolites were not measured in this study. Proteins that were identified in both Fe-replete and Fe-limited cultures are highlighted in grey. Proteins that were identified to be unique to the Fe-replete cultures are indicated in color. From top left – light blue: sugar and glycan biosynthesis, light purple: starch and sucrose metabolism (including photosynthesis, oxidative phosphorylation, carbon fixation), dark purple: glycolysis-gluconeogenesis (including TCA cycle), red: nucleotide metabolism, teal: lipid metabolism, orange: amino acid metabolism (including urea cycle).