Bivalve Proteomics
We are applying proteomics technology to better understand how bivalves interact with their natural and hatchery environments. These projects span species, life stages, and different proteomics technologies, and are described in detail below.
Our bivalve projects include:
Our bivalve projects include:
- Oyster physiology in response to ocean acidification
- Influences of the hatchery microbiome on oyster larvae
- Oyster larval response to temperature
- Proteomic biomarkers of geoduck maturation status
- Proteomic response of shellfish to environmental stress
|
The majority of this work is part of a collaboration with the Roberts Lab in the School of Aquatic and Fishery Sciences. The Roberts Lab focuses on the ecophysiology of aquatic organisms, from the nucleotide to the organismal level. |
Oyster physiology in response to ocean acidification
See our paper in BMC Genomics
Ocean acidification is a growing global threat to the marine environment. As land-based CO2 emissions increase, the increased atmospheric CO2 leads to greater CO2 concentrations in ocean surface water. Increased concentrations of CO2 in water throws the carbonate chemistry off balance and causes a decrease in ocean pH. This “acidification” has proven to be detrimental to many forms of marine life.
Our goal was to explore the susceptibility of Pacific oysters, Crassostrea gigas, to ocean acidification. Pacific oysters live in the nearshore environment where they are exposed to frequently changing environmental conditions, including changes in pH. Will oysters be negatively affected by future ocean acidification or are they pre-adapted to withstand these kinds of environmental changes? We exposed different groups of oysters to low or high pH water for 1 month and performed proteomics to uncover any potential effects of ocean acidification.
We found that ocean acidification does alter many important metabolic pathways in the oyster, but it did not affect mortality rate nor did it negatively impact all physiological parameters. However, ocean acidification did significantly alter the oysters’ proteomic response to a second stressor. By altering the oysters’ response to another stress, ocean acidification could impact how this species responds to the natural variability that occurs in its environment.
Our goal was to explore the susceptibility of Pacific oysters, Crassostrea gigas, to ocean acidification. Pacific oysters live in the nearshore environment where they are exposed to frequently changing environmental conditions, including changes in pH. Will oysters be negatively affected by future ocean acidification or are they pre-adapted to withstand these kinds of environmental changes? We exposed different groups of oysters to low or high pH water for 1 month and performed proteomics to uncover any potential effects of ocean acidification.
We found that ocean acidification does alter many important metabolic pathways in the oyster, but it did not affect mortality rate nor did it negatively impact all physiological parameters. However, ocean acidification did significantly alter the oysters’ proteomic response to a second stressor. By altering the oysters’ response to another stress, ocean acidification could impact how this species responds to the natural variability that occurs in its environment.
|
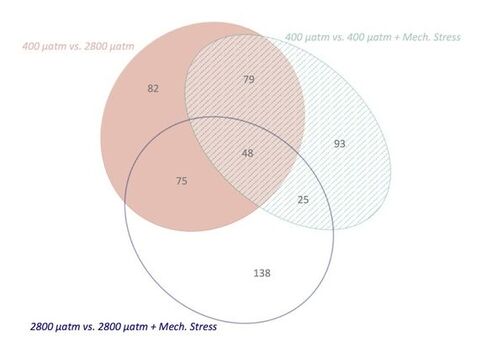
Venn diagram showing the overlapping and unique oyster proteins that respond to the environmental stresses of ocean acidification (solid pink), mechanical stress (aqua lines), and ocean acidification x mechanical stress (open blue). The gray numbers represent the number of differentially abundant proteins in each section of the ellipses. |
|
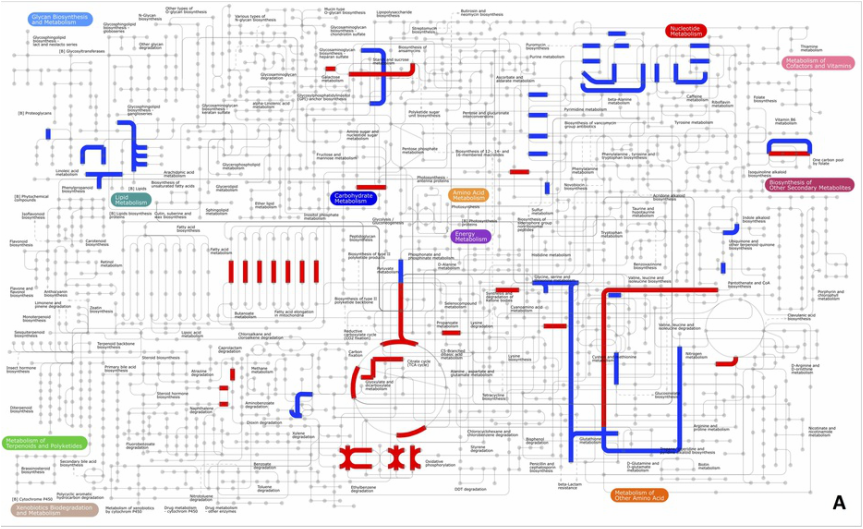
iPath output of differentially abundant proteins that changed levels in response to elevated pCO2 (2800 µatm versus 400 µatm). Blue lines represent pathways that are at lower abundance at high pCO2 and red lines represent pathways containing proteins of higher abundance. iPath (available free online) is one of the many visualization tools that we use to interpret patterns of protein abundance in our large datasets of hundreds to thousands of proteins. |
Influences of the hatchery microbiome on oyster larvae
One of the major bottlenecks in bivalve aquaculture is the unpredictable mass mortality events that occur during larval production. These mortality events occur after considerable resources have been used to raise a cohort of larvae. The causes of these events are rarely known and thus cannot be avoided in the future. We hypothesized that the intimate relationship between oyster larvae and their ambient microbiome may influence the success or failure of larval cohorts. We followed multiple cohorts of larvae at two different temperatures over the time period that usually includes the mass mortality events. We concurrently sampled the larvae and their rearing tank microbiome. We will explore the proteomic responses of both the larvae and their microbiome to find biomarkers of larval outcomes.
Oyster larval response to temperature
|
Larval rearing temperature can have a significant impact on oyster larval cohort success or failure. Higher temperatures can allow for faster larval growth, yielding cohorts that require fewer hatchery resources. However, mortality events can be common at high temperatures, making extended rearing at high temperatures relatively risky. In this experiment, Pacific oyster larvae were raised at 23°C and 29°C. Pooled larvae were analyzed using data independent acquisition (DIA) to achieve a deep sequencing of the proteome. This technique will allow us to detect proteins that occur at low abundances in the larval proteome. From these data, we will reconstruct the oyster larval physiological response to different hatchery rearing temperatures to reveal how that response may contribute to cohort success or failure.
|
|
Proteomic biomarkers of geoduck maturation status
|
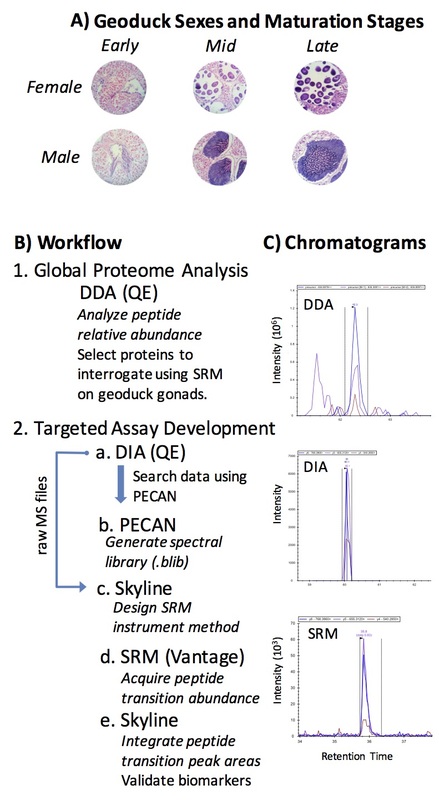
Geoduck clams are an increasingly important aquaculture product in the Pacific Northwest of the United States, while also holding essential ecological roles in the ecosystem. These long-lived clams are very fecund, but hatchery production of larvae is hindered by the inability to sex a live geoduck, thus resulting in asynchronous spawning of broodstock and unequal sex ratios. During geoduck reproductive maturation, the physiology of the gonad changes from sexually undifferentiated connective tissue to sex-specific, mature reproductive cells that can be released into the water column. We applied proteomics tools to uncover the cellular mechanisms underlying these physiological changes in the gonad. Sex- and stage-specific proteins were characterized using data dependent acquisition (DDA), or whole proteome profiling, on gonad tissue. Gonad proteomes became increasingly divergent between males and females as maturation progressed. The DDA data were leveraged to develop biomarkers of geoduck sex and maturation stage, analyzed with selected reaction monitoring (SRM) in gonad and hemolymph. The SRM assay yielded a reduced suite of peptides that can be used as an efficient assay to non-lethally determine geoduck sex and maturation stage pre-spawning. This is one of a few examples of cutting-edge proteomics being used to develop applicable tools for the aquaculture industry.
|
Proteomic response of shellfish to environmental stress
Naturally occurring shellfish populations occur across heterogeneous environments that offer differing degrees of protection from different environmental impacts. We outplanted Pacific oysters and geoduck in different pH environments throughout the Puget Sound region and then routinely sampled them to assess their physiological responses to their environment. We will use DIA methods to uncover their proteomic acclimations to each environment. This work is done in collaboration with the Department of Natural Resources.
Want to learn more about ocean acidification research at University of Washington? Check out the School of Aquatic and Fishery Sciences ocean acidification blog.